Automated technique helps identify cancer cell metabolism inhibitors
FINDINGS
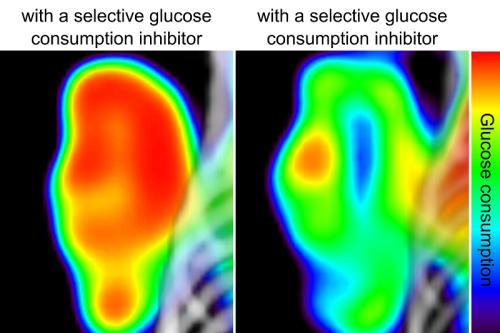
UCLA Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research investigators have developed a new automated method for testing hundreds of molecules at a time to find out which ones block cancer cells from consuming glucose — the sugars they need to spread and grow.
Using robotics, the researchers tested 3,555 compounds on non-small-cell lung cancer cells. They identified 97 molecules that inhibited these cancer cells from consuming glucose. In another study, these 97 molecules could be further tested to see which ones don’t interfere with healthy cell metabolism.
BACKGROUND
Cancer cells need to consume sugar in the form of glucose to grow and spread. Healthy cells also need sugar to carry out their normal cellular functions, such as creating energy for the body. Researchers have known for a long time that preventing cancer cells from consuming glucose will cause them to stop growing as quickly and make them easier to treat. But the challenge has been developing and identifying targeted therapies that are able to prevent cancer cells from consuming glucose while not interfering with the metabolism of healthy cells.
Even today, testing drugs that keep cancer cells from consuming glucose is a slow process because only one or two therapies are screened at a time. To change this, UCLA researchers have developed an automated technique using robotics that can test the effects of hundreds of molecules on non-small-cell lung cancer tumors at one time.
METHOD
With equipment in the UCLA Molecular Screening Shared Resource facility designed to test molecules against biological targets such as cancer cells, the researchers used a special glucose analogue that mimics naturally occurring glucose and rectangular plates with 384 individual wells for the test.
The researchers put non-small-cell lung cancer cells into each of the wells in the rectangular plates. They also put a different compound into each of the wells with the cancer cells. These compounds were given time to affect the cancer cells. After this, a glucose analogue was introduced into the cells, which the cells metabolized. The researchers treated the cells with a set of enzymes that use the metabolized glucose analogue to produce bioluminescence, or a light that can be tracked.
Using a plate reader that is able to track the amount of light that comes out of each well, the researchers recorded how much of the glucose molecule the tumors in each of the wells were able to metabolize.
IMPACT
UCLA’s new automated method will speed up how researchers test for therapies that block the metabolism of non-small-cell lung cancer tumors. In the future, this method may also be used to test for molecules that are able to inhibit the metabolism of other types of cancer cells.
AUTHORS
Authors include Chiara Ghezzi, Alicia Wong, Bao Ying Chen, Bernard Ribalet, Robert Damoiseaux and Peter Clark, who is the corresponding author. The authors have affiliations with UCLA departments and centers including: Crump Institute for Molecular Imaging, department of molecular and medical pharmacology, department of physiology, California NanoSystems Institute, Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research, and the UCLA Jonsson Comprehensive Cancer Center.
JOURNAL
The research was published in Nature Communications.
FUNDING
This study was in part funded by the National Institutes of Health.
